What Determines The Shape Of A Protein
listenit
Mar 18, 2025 · 5 min read
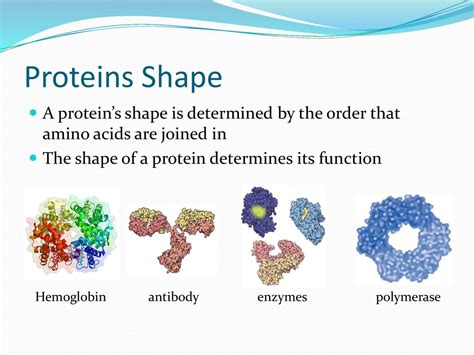
Table of Contents
What Determines the Shape of a Protein?
Proteins are the workhorses of the cell, carrying out a vast array of functions essential for life. Their remarkable versatility stems from their incredibly diverse shapes, which are intricately linked to their specific functions. But what exactly determines the precise three-dimensional structure of a protein? The answer is a complex interplay of several factors, starting from the primary sequence of amino acids and extending to the influence of the cellular environment.
The Primary Structure: The Blueprint of Protein Shape
The fundamental determinant of a protein's shape is its primary structure: the linear sequence of amino acids. This sequence is dictated by the genetic code, specifically the order of nucleotides in the corresponding gene. Each amino acid possesses unique chemical properties, including its size, charge, hydrophobicity (water-loving or water-fearing), and ability to form hydrogen bonds. These properties are crucial in shaping the higher-order structures.
The Role of Amino Acid Properties
Consider the different amino acid side chains:
- Hydrophobic amino acids: These tend to cluster together in the protein's interior, away from the surrounding aqueous environment. This hydrophobic interaction is a significant driving force in protein folding.
- Hydrophilic amino acids: These are often found on the protein's surface, interacting with water molecules.
- Charged amino acids: These can form electrostatic interactions (ionic bonds) with each other or with other charged molecules in the cell.
- Amino acids capable of forming disulfide bonds: Cysteine residues can form strong covalent bonds (disulfide bridges) that stabilize the protein's structure.
The precise arrangement of these amino acids along the polypeptide chain dictates how the protein will fold. A single amino acid substitution can dramatically alter the protein's final shape and, consequently, its function. This is exemplified by diseases like sickle cell anemia, where a single amino acid change in hemoglobin leads to a drastically altered protein shape and impaired oxygen transport.
Secondary Structure: Local Folding Patterns
As the polypeptide chain emerges from the ribosome, it begins to fold into local structures called secondary structures. These are stabilized primarily by hydrogen bonds between the backbone atoms of the amino acids. The two most common secondary structures are:
Alpha-Helices:
Imagine a coiled spring. This is analogous to an alpha-helix, where the polypeptide chain forms a right-handed helix stabilized by hydrogen bonds between the carbonyl oxygen of one amino acid and the amide hydrogen of an amino acid four residues down the chain. The presence of certain amino acids, like proline (which disrupts the helix), can influence the formation and stability of alpha-helices.
Beta-Sheets:
In contrast to the helical alpha-structure, beta-sheets consist of extended polypeptide chains arranged side-by-side, forming a sheet-like structure. These strands are stabilized by hydrogen bonds between adjacent strands. Beta-sheets can be parallel (strands run in the same direction) or antiparallel (strands run in opposite directions). The arrangement of strands contributes to the overall shape and stability of the beta-sheet.
Tertiary Structure: The 3D Puzzle
The tertiary structure represents the overall three-dimensional arrangement of a single polypeptide chain. This structure is determined by a complex interplay of various forces, including:
- Hydrophobic interactions: As mentioned earlier, hydrophobic amino acids cluster together in the protein's core, minimizing their contact with water.
- Hydrogen bonds: These weaker bonds contribute to the stability of the tertiary structure by forming between various side chains and the polypeptide backbone.
- Ionic interactions (salt bridges): Attractive forces between oppositely charged amino acid side chains stabilize the protein's structure.
- Disulfide bonds: These strong covalent bonds between cysteine residues significantly contribute to the stability of the tertiary structure, particularly in extracellular proteins.
- van der Waals forces: These weak, short-range interactions between atoms contribute to the overall packing and stability of the protein.
The tertiary structure is often described using different motifs, such as:
- Domains: These are independently folded units within a protein, often with specific functions. A single protein can consist of multiple domains, each contributing to the overall function.
- Loops and turns: These regions connect secondary structure elements and play critical roles in protein function and flexibility.
Quaternary Structure: The Protein Complex
Some proteins consist of multiple polypeptide chains, each with its own tertiary structure, assembled into a larger complex. This is called the quaternary structure. The interaction between these subunits is governed by the same forces that stabilize the tertiary structure, including hydrophobic interactions, hydrogen bonds, ionic interactions, and disulfide bonds. Examples of proteins with quaternary structure include hemoglobin (with four subunits) and many enzymes.
The Influence of the Cellular Environment
The cellular environment significantly influences protein folding. Factors such as:
- Molecular chaperones: These proteins assist in the proper folding of other proteins, preventing aggregation and misfolding. They can bind to unfolded or partially folded proteins, providing a protected environment for proper folding.
- pH: Changes in pH can alter the charge of amino acid side chains, affecting electrostatic interactions and potentially disrupting the protein's structure.
- Temperature: Extreme temperatures can disrupt the weak interactions that maintain the protein's structure, leading to denaturation (unfolding).
- Presence of other molecules: The presence of ligands (molecules that bind to proteins) can induce conformational changes, altering the protein's shape and function.
Protein Folding Prediction and Misfolding
Predicting the three-dimensional structure of a protein solely from its amino acid sequence remains a significant challenge in computational biology. While significant progress has been made, accurately predicting the precise shape of a protein is still not fully solved. This difficulty arises from the vast number of possible conformations a protein can adopt and the complex interplay of various forces influencing its folding.
Protein misfolding can have dire consequences, leading to the formation of aggregates that are associated with various diseases, including Alzheimer's disease, Parkinson's disease, and cystic fibrosis. These aggregates can interfere with cellular function and cause cell death.
Conclusion: A Multifaceted Process
Determining the shape of a protein is a complex and multifaceted process, governed by a intricate interplay of primary sequence, local folding patterns (secondary structure), overall three-dimensional arrangement (tertiary structure), potential multi-subunit assemblies (quaternary structure), and the cellular environment. Understanding these factors is crucial not only for comprehending the fundamental principles of biology but also for developing therapeutic strategies to address protein misfolding diseases and to design novel proteins with desired properties. The ongoing research in protein folding continues to unravel the secrets of these remarkable biological molecules and their essential roles in life.
Latest Posts
Latest Posts
-
Why Are Alkyl Groups Electron Donating
Mar 18, 2025
-
What Is The Longest Phase In Mitosis
Mar 18, 2025
-
How Many Quarts Are In 7 Pints
Mar 18, 2025
-
Is Speed A Vector Or A Scalar
Mar 18, 2025
-
What Is The Valence Electrons Of Nitrogen
Mar 18, 2025
Related Post
Thank you for visiting our website which covers about What Determines The Shape Of A Protein . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.
